Research
The role of riboregulators and RNA-binding proteins in regulating adaptive responses in microorganisms
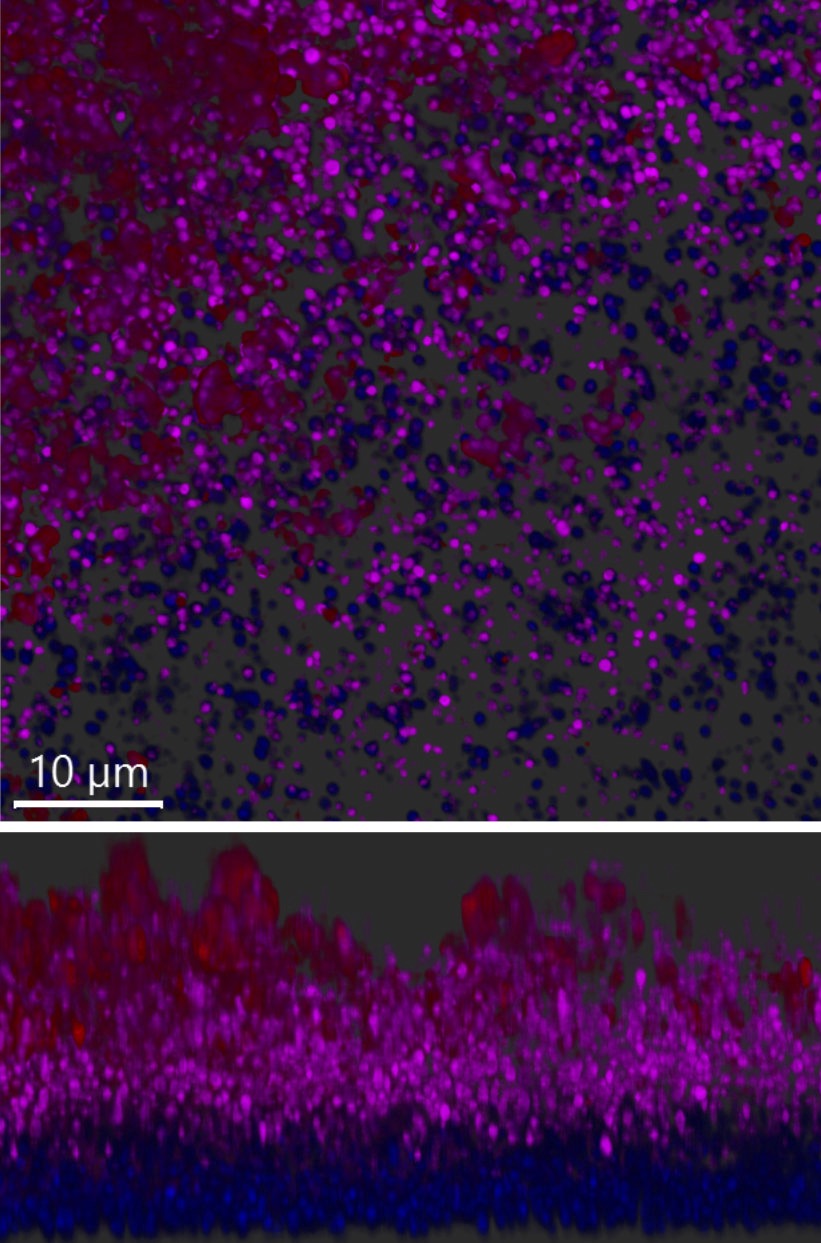
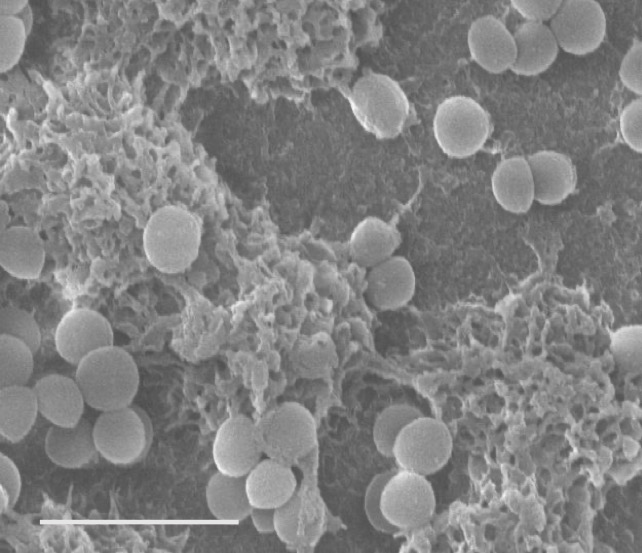
Figure legend: Left image: S. aureus microscopy image. Second image: S. aureus biofilms stained with Z and B-DNA antibodies and imaged using a confocal microscope. Right image: Cryo-SEM image of an S. aureus biofilm grown on catheters. Confocal and cryo-SEM images were generated by Mehak Chauhan.
The Granneman lab is studying the role of riboregulators and RNA binding proteins in regulation of gene expression during rapid adaptive responses, for example, when cells are deprived of essential nutrients or enter a hostile host environment.
Many microorganisms, in particular human pathogens, have evolved clever mechanisms to be able to very rapidly adapt to stress caused by environmental changes. This enables them to efficiently maintain cellular homeostasis even in hostile environments. Our goal is to gain mechanistic insights into regulatory strategies used by these organisms to adapt to stress.
Our research focusses on riboregulators and RNA-binding proteins that play a key role in stress adaptation. We use the yeast Saccharomyces cerevisiae and multi-drug resistant bacteria as model organisms. We use highly interdisciplinary approaches to address our research questions, including protein and RNA biochemical techniques, structural biology, bioinformatics, high-throughput sequencing, proteomics and cell biology (eg. see van Nues et al 2017, Iosub et al 2019 and Chu et al, 2022).
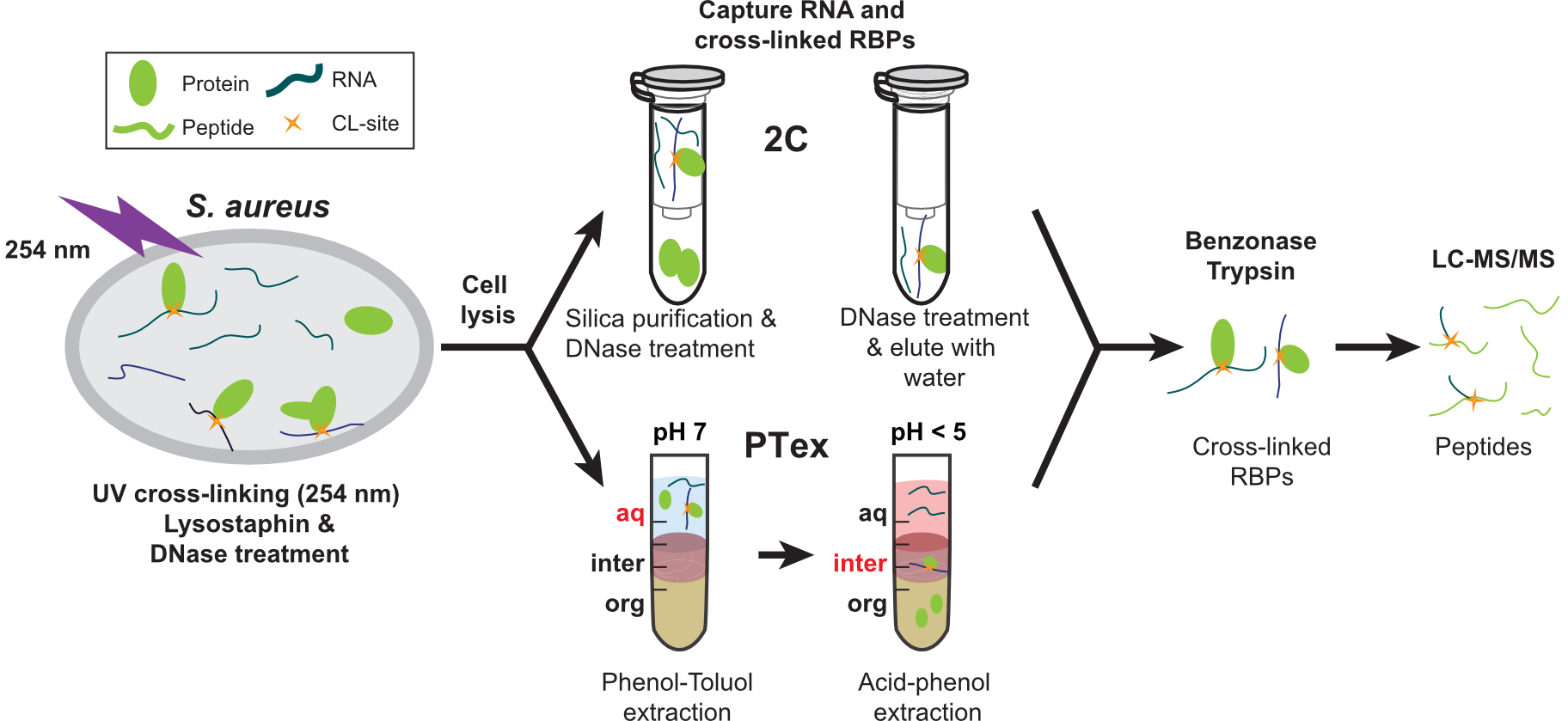
Very recently, we unearthed the RNA-binding proteome (Chu et al, 2022) and sRNA-RNA interactomes (McKellar et al, 2022) in methicillin-resistant Staphylococcus aureus. These studies uncovered many unusual RNA-binding proteins and sRNA-RNA interactions involved in controlling virulence and antibiotic resistance.
In the yeast Saccharomyces cerevisiae we recently discovered a new mechanism that reduces gene expression noise that involves co-transcriptional decay of mRNA (Esteban-Serna et al., 2025).
We also developed a new and simple method for conditional depletion of proteins from the nucleus that is non-toxic and fully reversible (Esteban-Serna et al., 2025b).
This work is sponsored by generous contributions from:



