Hardware
Here we showcase the hardware we developed in the Granneman lab for rapid UV cross-linking of protein-RNA complexes in living cells. We worked closely with UVO3, a UK based company, to develop these tools and bring them to the market.
Since 2013 we have been working closely with UVO3, a UK based company, to develop new tools for rapid UV protein-RNA cross-linking in vivo and cell harvesting. We brought it on the market in 2018 and it has already been sold to labs all over the world.
The first UV cross-linker: The Megatron
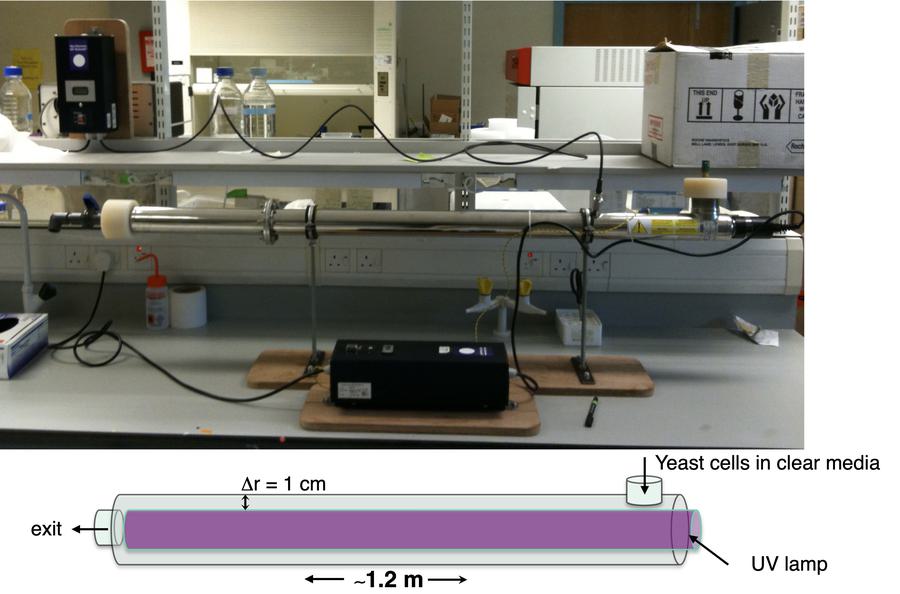
The first UV cross-linker we got from UVO3 was simply one of their water sterilisation units that we modified to UV cross-link yeast cultures. This is showing the unit from the side. It really wasn't a spectacular thing but simplicity was key here and it worked. This unit was published in the 2011 EMBO journal paper and several labs have since gotten one too.
Vari-X-linker UV cross-linker

Information about the Vari-X-linker UV cross-linker

We then decided to take things a step further with UVO3. This resulted in the very first iteration of the Vari-X-Linker, which we developed together with Peter Wadsworth and Andrew Langford. We still have it (!) and it works like a charm. We used this machine for the van Nues et al 2017 publication.
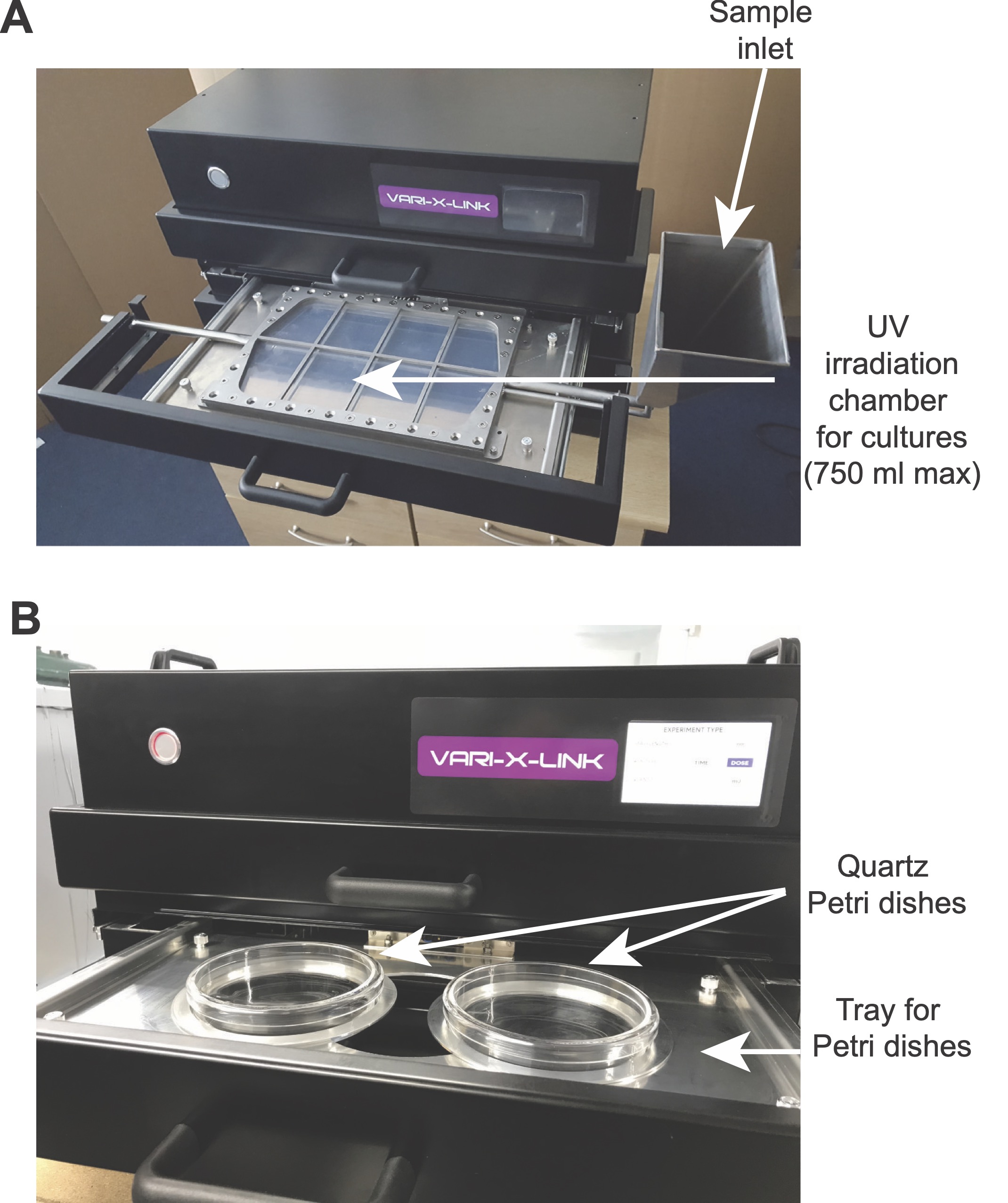
Finally, UVO3 produced the Mark I, the very first Vari-X-Linker that was made commercially available. This was the very first high intensity UV cross-linker on the market. It allows you to UV cross-link cells that are actively growing, which enabled us to capture more transient interactions and the whole cross-linking process is also much faster. The quartz petridishes were made to UV cross-link smaller volumes of cells, such as mammalian cells. This machine is described in our JoVe manuscript that outlines our time-resolved CRAC protocol for yeast and mammalian cells.

This is Rob van Nues explaining to me how this new kit works.

We then added rapid filtration units that UVO3 made for us (can also be purchased).
Time-resolved CRAC/CLIP
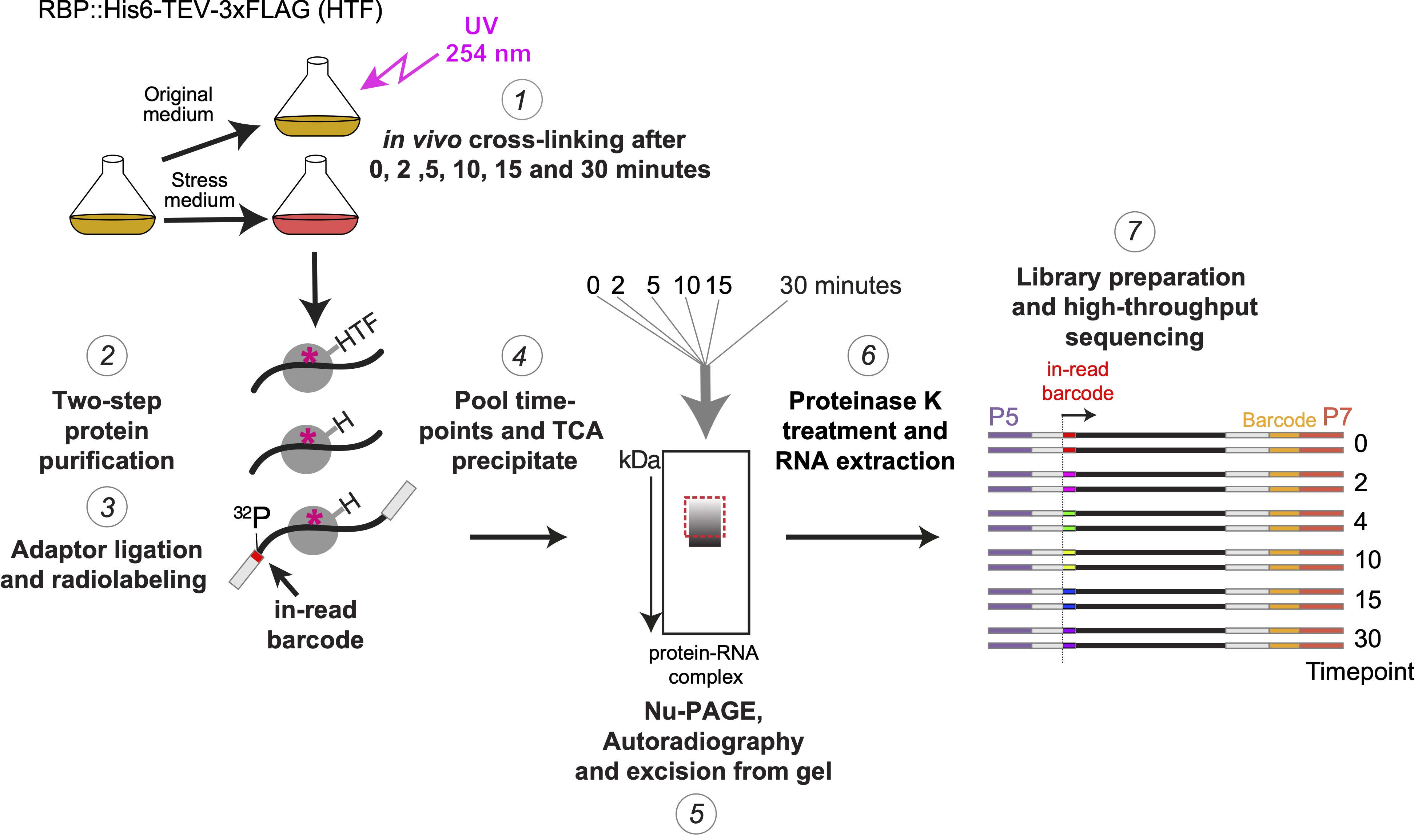
How do we use the Vari-X-linker and these filtration devices?
We pioneered time-resolved CLIP analyses and we used it to monitor interactions between proteins and their RNA substrates at minute time-point resolution after cells are shifted to a stress-inducing medium. We use in-read barcodes for each CRAC experiment and all barcoded RNAs are pooled once the complexes are eluted from the beads. We are also working on adding spike-ins to the library preparation method.